Explainer: DNA hunters
Genetic clues can tell scientists which organisms have been moving through the environment

Scientists can extract DNA (parts of which are displayed on screen) from samples they picked up in the environment. They then look for patterns in this genetic material to identify which species had left it. In this way, biologists can “detect” what organisms had been around.
Wavebreakmedia/iStockphoto
“Traces of DNA are left behind by every species everywhere,” says Ryan Kelly. He is an ecologist with the University of Washington at Seattle. He also works at the Center for Ocean Solutions at Stanford University in Palo Alto, Calif. Scratch an itch, he says, and you shed skin cells containing your DNA. Pets and other animals leave behind bits of dead skin known as dander. Reptiles shed skin as they grow. There’s even DNA in poop.
“Just like forensic scientists do at a crime scene every day, we are detecting that trail of DNA that’s left behind,” explains David Lodge. He’s a biologist at the University of Notre Dame in South Bend, Ind. And he hunts for signs an animal has been around by scouting for bits of the DNA it had shed. Scientists refer to this genetic litter as e-DNA. Here, the “e” stands for environmental.
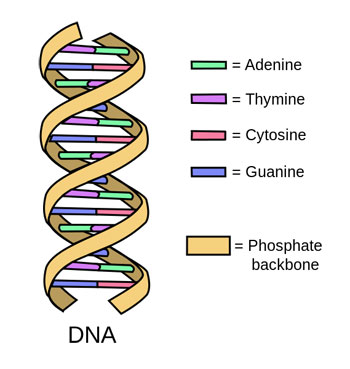
DNA is the genetic material found in the cells of all living organisms. It looks like a long, twisting ladder. Each rung on that ladder is a pair of chemicals called nucleotides. There are four of these chemicals: adenine, thymine, cytosine and guanine. Scientists refer to them as A, T, C and G, for short. Each nucleotide on one long side of the ladder must pair with a specific one on the other side. A’s only pair with T’s. Any C’s must pair with G’s.
Humans have about 3 billion rungs, or base pairs, in their DNA. Other species have more or fewer. The order of base pairs in every individual’s DNA is unique, but its pattern will be very similar to that of other members of its species. Scientists can use that code to identify a species.
Scientists prowl for eDNA throughout the environment, in water, soil, ice cores — even the guts of leeches.
Researchers in the lab start with some environmental sample. It might be a leaf, a vial of water, some animal tissue or dust. The first step is to isolate cells of one or more living organisms in the sample. Then they add these to some hard-working chemicals. One chemical breaks down cell walls to release their DNA. Another chemical grabs onto proteins and other materials that are not DNA. A machine then spins the mix at high speed. DNA-filled liquid floats to the top. Everything else sinks to the bottom. More steps get rid of extra liquid from that top layer.
Additional work might be needed along the way to “clean up” samples even more, explains Margaret Hunter. She’s a geneticist with the U.S. Geological Survey in Gainesville, Fla. For example, her lab had to remove certain compounds produced by local trees. She was looking for DNA from invasive Burmese pythons. Those tree chemicals could have stopped reactions that should take place in the next set of steps in her search for python eDNA.
In projects focused on some particular species, such as the python, those next steps search through the sample for a particular span of rungs from the DNA’s ladder-like structure. This specific span of the DNA molecule is known as a sequence. And it will be unique to its species.
If it’s found, the method will then make many, many copies of that DNA fragment. This copying process is called the polymerase (Puh-LIM-er-ase) chain reaction, or PCR. In effect, it works like a photocopier for DNA.
Using PCR will boost that DNA signal, explains Eva Egelyng Sigsgaard. She’s a biologist with the Natural History Museum of Denmark in Copenhagen. Think of turning up the volume on your cell phone. After you amplify the sound, you can hear the ring over any background noise in a room. Similarly, she says, “When we amplify the DNA, we make large amounts of the specific DNA that we’re looking for.” That makes this DNA stand out against the “noise” of other species’ genetic debris.
Hunter at USGS tailored her work to find and copy a part of the Burmese python’s DNA that is different from that in all other species. In 2015, that let her team show for the first time that the big snakes could be detected through eDNA. If eDNA sampling becomes more routine, it could help scientists track — or scout for — evidence of rare or camouflaged species at relatively low cost.







