New tools can fix genes one letter at a time
These 'editors' can fix DNA mistakes that may underlie disease
Scientists have used two new versions of CRISPR technology to edit DNA one letter at a time. The new tools change bits of DNA like a pencil, instead of cutting it like scissors.
Wildpixel/iStockphoto
There’s a fairly simple alphabet that spells out the directions for what genes want the body’s cells to do. But sometimes there’s a mistake — essentially a typo — in those directions. Now, working somewhat like a pencil, new tools can edit the body’s genes one letter at a time. They can fix about half of the simple typos that cause human diseases.
The new tools are based on a technology called CRISPR/Cas9. It’s like a set of molecular scissors that snips DNA. Scientists can guide the scissors to the exact spot in an organism’s genetic instruction book where they want to cut something out. They’ve used the tool to make faulty genes — mutations — or correct them. And they’ve done this in animals and in human cells.
Scientists also have ways to use CRISPR/Cas9 to change gene instructions without cutting DNA at all. These tools are called “base editors.” A DNA base is a molecule represented by a single letter in the body’s genetic code. There are four DNA bases, known as A, C, T and G.
Base editors can fix one-letter typos in the genetic code. So far, they’ve only been able to fix one type of error: They’ve turned C’s into T’s.
Using these tools, scientists have changed genes in plants, fish, mice — even in human embryos.
Editing DNA in this way may be safer than cutting it, says Gene Yeo. He’s a biologist at the University of California, San Diego. “We know there are drawbacks to cutting DNA,” he says. After CRISPR breaks DNA, a cell’s own machinery pastes it back together. But mistakes often occur during this process. And CRISPR sometimes makes extra DNA cuts at places similar to the target. That means it can create new mutations by accident. Such “permanent irreversible edits at the wrong place in the DNA could be bad,” Yeo says.
Two papers now describe different ways to solve that problem.
David Liu and colleagues tweaked the CRISPR/Cas9 gene editor so that it turns the DNA base adenine (A) into guanine (G). They shared their work October 25 in Nature.
In a second study, Feng Zhang and colleagues redesiged a gene editor called CRISPR/Cas13 to correct the same typos in RNA. RNA is a molecule similar to DNA. RNA carries messages inside cells. This study was also published October 25, but in the journal Science.
The new tools expand the toolkit scientists have for correcting diseases. Now researchers can rewrite all four letters in the genetic code.
Repairing rungs
DNA has two strands that give it the shaped of a long, twisted ladder. Bases on one side of the ladder always pair up with certain bases on the opposite side. The molecule adenine (A) always pairs with thymine (T). Guanine (G) always pairs with cytosine (C). These base pairs form the ladder’s rungs.
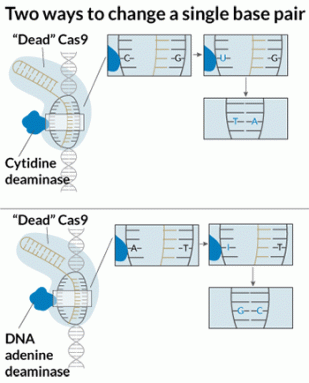
Mutations that change C-G base pairs to T-A pairs happen all the time. Some 100 to 500 of these changes occur every day in everyone’s body. Most of those mutations are likely harmless. But some can cause serious changes to a gene’s instructions — changes that lead to disease.
Liu is a biological chemist at Harvard University in Cambridge, Mass. Scientists have linked some 32,000 DNA mutations to genetic diseases in people, he notes. And about half are the C-G to T-A swaps. Until now, he says, there was little anyone could do about them.
RNA is the chemical cousin of DNA. It uses nearly the same code to spell out genetic information as DNA does. But instead of the base thymine (T), RNA uses uracil (U). In RNA, some naturally occurring enzymes can reverse the mutations that turn C-G pairs into T-A pairs. These enzymes chemically turn adenine (A) into another molecule called inosine (I). The cell reads this I as a G. Such RNA editing happens frequently, and naturally, in octopuses and other cephalopods. Sometimes it also happens in people.
Zhang and colleagues made one of these RNA-editing enzymes into a programmable gene-editing tool. Zhang is a scientist at the Broad Institute of MIT and Harvard in Cambridge, Mass. He also was one of the first developers of CRISPR.
To create their new tool, these researchers started with CRISPR/Cas13. Normally, this tool works like a molecular scissors that cuts RNA. The researchers tweaked the tool so that it grasped RNA instead of slicing it. Then they attached the part of the RNA enzyme that converts A to I.
The researchers called the new creation REPAIR. They tested it on human cells growing in dishes. The tool edited up to about 27 percent of the RNAs of two genes. The researchers did not find any unwanted changes.
Permanent fixes
Editing RNA is good for temporary fixes, such as turning off proteins that cause inflammation. But many mutations need permanent DNA fixes, notes Liu.
Last year, his team made a base editor that converts C to T. Other researchers have used it to fix a blood disease in human embryos. But that tool couldn’t make the opposite change, switching an A to a G.
Unlike with RNA, there’s no enzyme that naturally changes A’s to G’s in DNA. So Nicole Gaudelli forced E. coli bacteria to evolve one. She works in Liu’s lab.
First Gaudelli engineered bacteria to have a mutation in a gene that helps them fight an antibiotic. Now when she fed the bacteria that antibiotic, it killed them. Only by correcting the mutation, switching an A to G, would the bacteria live. It took lots of work, but eventually the bacteria succeeded.
The researchers called this new DNA-base converter “TadA.” Gaudelli attached it to a “dead” version of Cas9, one that couldn’t cut both strands of DNA. The result was a base editor that could switch A-T base pairs into G-C pairs. The scientists named their new tool ABE. In tests on human cells, it worked in about half of all cells tested.
This base editor works more like a pencil than scissors, says Liu. In lab dishes of human cells, his team corrected a mutation. The cells came from a patient with a disorder that affects how the blood stores iron. In other cells, the team re-created helpful mutations that protect against a disease called sickle cell anemia.
Such base editing seems safer than traditional cut-and-paste editing, other researchers now report in the October issue of Protein & Cell.
Liu’s results seem to agree. His team found that cut-and-paste CRISPR/Cas9 edits made changes at nine of 12 possible “off-target” sites. This happened about 14 percent of the time. But the new A-to-G base editor changed just four of the 12 off-target sites. And those errors happened only 1.3 percent of the time.
That’s not to say cut-and-paste editing isn’t useful, Liu says. “Sometimes, if your task is to cut something, you’re not going to do that with a pencil. You need scissors.”
Editor’s note: Feng Zhang is on the board of trustees of the Society for Science & the Public, which publishes Science News for Students.







