Three take home chemistry Nobel for harnessing protein ‘evolution’
The scientists created bio-based techniques to speed up the development of drugs, fuels and more

This year’s Nobel Prize in chemistry went to Frances Arnold (left), George P. Smith (middle) and Gregory P. Winter (right) for research on proteins, including enzymes and antibodies.
CalTech; University of Missouri; Aga Machaj/Wikimedia Commons (CC BY-SA 4.0)
By Maria Temming and Laurel Hamers
Biologists often refer to evolution as the way living things adapt, genetically, to their environment. But scientists sometimes harness “evolution” to help out in other fields of science. Three who did were today awarded the 2018 Nobel Prize in chemistry. They developed ways to build proteins in the lab through the use of evolution.
The proteins that can be fashioned this way are suitable for a broad range of uses. These range from new drugs to biofuels.
Frances Arnold works at the California Institute of Technology in Pasadena. She will take home half of this year’s prize of 9 million Swedish kronor (about $1 million). She worked on enzymes. These are materials that cells create to speed up chemical reactions. Arnold learned how to make new enzymes from old ones. These new enzymes can be used to make biofuels, environmentally friendly detergents and other products.
Arnold is only the fifth woman over the past 117 years to win a Nobel Prize in chemistry.
Gregory Winter works in England at the University of Cambridge. George Smith is at the University of Missouri in Columbia. They will split the other half of this year’s chemistry prize. They found a way to build peptides (short strings of amino acids) using viruses. Some of the peptides are used to make antibodies and other medicines.
“Wow, well-deserved!” says Paul Dalby of this year’s prize winners. “Protein engineering as a field is absolutely founded upon their work,” he notes. Dalby is a biochemical engineer. He works in England at University College London.
All three winners will collect a medal and their shares of the prize money at a ceremony in Stockholm, Sweden on December 10. That’s the date in 1896 on which Alfred Nobel died.
Mining the chemical know-how of living organisms
In the late 1980s, Arnold wanted to make an enzyme to break down a milk protein, casein, in a solvent other than water. She could try to manually link up the amino-acid building blocks of that enzyme to give it the right properties. That would require knowing which amino acids to change. Instead, she opted for a more hands-off approach.
Arnold’s insight, says Jesse Bloom, “was to recognize that the most amazing molecules in the world weren’t created by chemists.” He is a microbiologist at the Fred Hutchinson Cancer Research Center in Seattle, Wash. Those cool molecules are made in the cells of living things. What’s more, he notes, “Biology didn’t make these chemicals using the methods we might learn in an organic chemistry class. Rather, it worked by evolution.”
Arnold first made many copies of the DNA that contains instructions for making the original enzyme. She put that DNA into bacteria. As the bacteria grew, genetic differences — changes known as mutations — arose by accident.
Each batch of bacteria could have many different versions of the mutant enzyme-making genes. The bacteria also acted like living factories. They churned out many copies of each mutant enzyme. Arnold picked the version of that enzyme that did the best job at breaking down casein in a particular solvent. Then she repeated the mutation process, starting the best-working enzyme.
Her bacteria went through rounds of mutation-making. Each time she selected the best-working enzyme that some new mutant bacterium had made. In the end, Arnold was left with an extremely efficient, custom-made molecule. The technique she used is now known as “directed evolution.” It allows scientists to fine-tune the chemistry of their proteins in much the same way that Mother Nature does it. The difference: Arnold’s technique is thousands of times faster.
Researchers have used directed evolution to create enzymes that jump-start chemical reactions for creating new drugs and eco-friendly biofuels. Arnold’s lab has used this process to also create enzymes that help forge chemical connections not found in nature.
(story continues below image)
Going viral
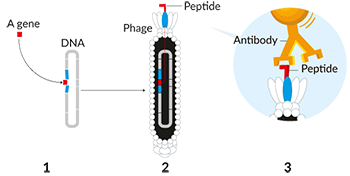
The other half of the Nobel prize honors work on a procedure to make chemicals using viruses. Certain viruses kill bacteria. Those viruses are known as phages (FAYZH-es). That’s short for bacteriophages. These are simple microbes. They’re just bundles of genetic material enclosed inside protein shells.
Smith and Winter created phage programmed with genes to build various proteins (or enzymes). Then they injected the phage into bacteria and let the phage reproduce inside them. Sometimes they mutated, making new versions of the target proteins. Those proteins would show up on the phage’s outer shells, earning the name “phage display.”
In the 1980s, Smith started work on this process by inserting certain genes into the DNA of phages. Those genes told cells how to make various proteins. Their gene-tweaking created phages that displayed the target proteins on their surface.
Smith’s goal had been to figure out which genes created which proteins. He essentially went fishing around a phage soup with a molecule known to hook a particular protein. This let him to pull out only the phages armed with that particular surface protein.
In 1985, Smith created a phage that held on its surface a piece of protein known as a peptide. He then used a peptide-binding chemical to glom onto this phage and pull it out of the crowd.
Other researchers have gone on to use his method to breed phage that wear antibodies and other biological molecules on their surfaces.
(story continues below image)
On to antibodies
The human body naturally makes hundreds of thousands of antibodies. They are designed to latch onto viruses and bacteria. Their presence puts a bullseye on these microbes, marking them for destruction by immune cells. But researchers had long wanted to create antibodies in the lab to tackle various diseases. These could work as new drugs. In 1990, Winter used his phage-display method to create a phage armed with part of an antibody.
The antibody he used binds to a molecule known as phOx. Winter then used phOx to latch onto a phage wearing that antibody. It pulled the right phage out of a soup of four million other types of phage.
Winter then started using directed evolution, much as Arnold had been. He first created a pool of phage whose genes programmed them to produce billions of different antibodies. From that group, Winter now used a target molecule like phOx to serve as a hook. It would collect only those phage that were best at binding to it. From those phage, Winter bred a whole new generation of antibody-bearing phage. Then again, he used the target molecule to pick out the best of this newer bunch.
Using this survival-of-the-fittest-phage technique, Winter’s team created a drug based on an antibody. It treats the source of inflammation in people with autoimmune diseases. Such ailments hijack the immune system to attack a patient’s body. In 2002, a drug made this way, called Humira, was cleared to treat rheumatoid arthritis (RU-muh-toid Arth-RY-tis). Today, doctors use this drug to also treat two other autoimmune diseases.
Phage display “is an extremely versatile technology,” says Jonathan Lai. He’s a biochemist at the Albert Einstein College of Medicine in New York City. His lab uses the technique to make new vaccines. Other researchers have used phage display to produce antibodies that fight cancer or treat lupus (LOO-puhs).
Jon Lorsch directs the National Institute of General Medical Sciences in Bethesda, Md. He says that the discoveries by this year’s Nobel winners are “a great demonstration of how studying fundamental biological questions like the natural process of evolution can lead to great breakthroughs in technology and medicine.”







